Recently, Prof. Zhi John Lu’s research group from the School of Life Sciences of Tsinghua University, and collaborators, published a research paper titled "RNA-ligand interaction scoring via data perturbation and augmentation modeling" in Nature Computational Science. This study breaks through the limitation of traditional drug design methods that rely on three-dimensional structures. In view of the lack of RNA structures, a sequence-based input RNA-small molecule interaction AI prediction model, RNAsmol, is proposed. This model provides an efficient computational tool for the development of small molecule drugs targeting RNA, and provides new solutions and ideas for AI-assisted drug design that does not rely on three-dimensional structures.
Author's introduction:
" RNA, not DNA, is the computational engine of the cell."
In our small molecule drug prediction work (RNAmol) and previous small RNA drug prediction work (OligoFormer), we tried to use a simple, RNA-specific grammar (such as A-U, G-C, G-U) to represent RNA molecules, without using a three-dimensional structural model or an all-atom model in the physical sense. This specific and simple grammar not only achieved unexpected results in the above work, but also made us more convinced of an academic hypothesis that is familiar in the RNA field: the origin of life is a RNA world. This seemingly simple RNA language may have the basic elements of the origin of life or even the origin of the universe: information replication, transmission and mutation. We hope that our attempts and practices will not only inspire the field of drug design, but also serve as a starting point for research in other directions of life science and computational science.
Background
Currently, the vast majority of clinical drugs target proteins. However, many proteins are often considered "difficult to drug" or "undruggable" due to the lack of suitable structural pockets. Of the approximately 20,000 protein-coding genes in human (accounting for about 1.5% of the total length of the human genome), about 10%-15% are directly related to disease; and of these genes, it is estimated that only 700-900 protein products are druggable (accounting for only about 0.05% of the total length of the human genome). On the other hand, about 70% or more of the human genome is transcribed into RNA, most of which is noncoding RNA (ncRNA). Therefore, in recent years, more and more researchers have begun to try to use RNA as a drug target and have initially demonstrated the feasibility of this strategy. The cost of new drug development is expensive and the cycle is long. The use of computer-aided drug design can greatly reduce the cost of research and development, and assist and accelerate the development of small molecule drugs targeting RNA. However, due to the lack of public RNA-small molecule interactions and known high-resolution RNA structure data, the development of data-driven deep learning models still faces many challenges.
Research Content
Zhi John Lu's laboratory has long been committed to RNA bioinformatics research and has accumulated a lot of scientific research experience in the computational design of RNA-siRNA/shRNA, RNA-protein, and RNA-ligand. In this latest work, the authors used data perturbation and augmentation strategies to develop a deep learning model for RNA-small molecule binding and built an AI prediction method RNAsmol for RNA-small molecule interaction scoring. Compared with other computational methods, RNAsmol not only has better prediction performance, but also has the potential to be widely used in a variety of drug screening scenarios. It can still predict small molecule drugs for many RNA molecules without three-dimensional structure information.
1. RNAsmol, a deep learning framework based on data perturbation and augmentation
The RNAsmol framework proposed in this work is a deep learning method that combines data perturbation and data augmentation strategies. In this framework, data perturbation simulates the data diversity in the real environment by randomly perturbing the training data, thereby helping the model to better learn the rules of RNA-small molecule binding. Data augmentation increases the model's ability to identify unknown spaces by generating virtual negative samples and potential unlabeled samples based on known interactions. This strategy not only improves the robustness of the model, but also helps it better capture different types of interaction patterns. In addition, the model combines a graph-based molecular feature representation method and a graph diffusion convolution module to model the structure of drug small molecules, and uses a feature fusion module based on an attention mechanism to weightedly integrate target and drug molecule features in multiple modalities, ultimately achieving the scoring prediction between RNA targets and small molecules.
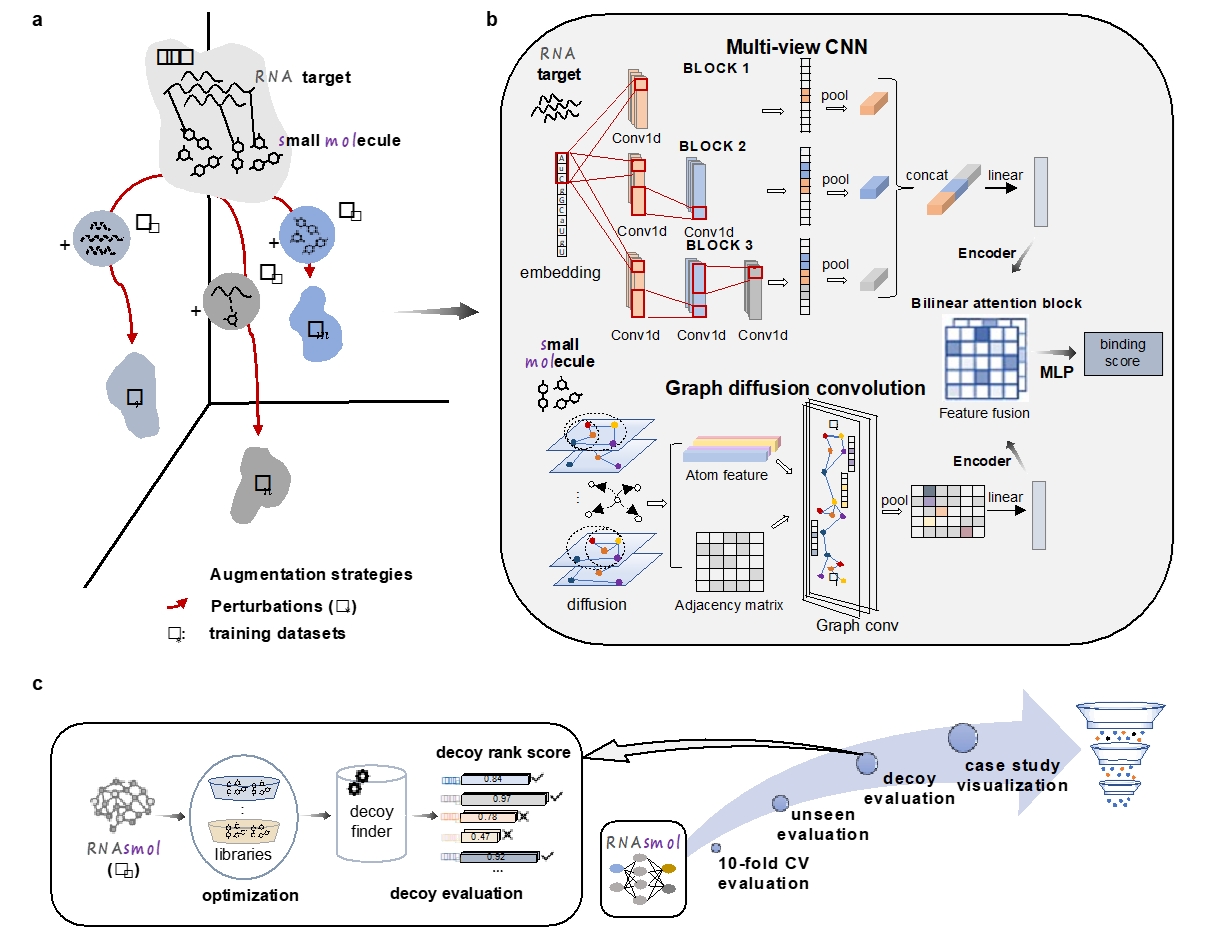
Figure 1. RNAsmol model and overall computational framework
2. RNAsmol can accurately classify RNA-small molecule interactions in data perturbation space
In data perturbation space, RNAsmol effectively reduces the deviation between real negative samples and unknown interaction space through a perturbation strategy. This strategy generates potential "negative" samples by perturbing known negative samples, and at the same time expands the boundaries of known positive and negative samples through data augmentation techniques. This enables the model to better understand the binding rules between RNA and small molecules, especially in the case of unbalanced data, avoiding the model's bias towards known positive and negative samples. Experimental results show that RNAsmol outperforms traditional methods in 10-fold cross validation, with the average AUROC (area under the curve) index improved by about 8%, and the performance improved by about 16% in the evaluation of unseen samples. This advantage proves the effectiveness of this method in sparse data scenarios and further promotes computational research on RNA-small molecule binding prediction.
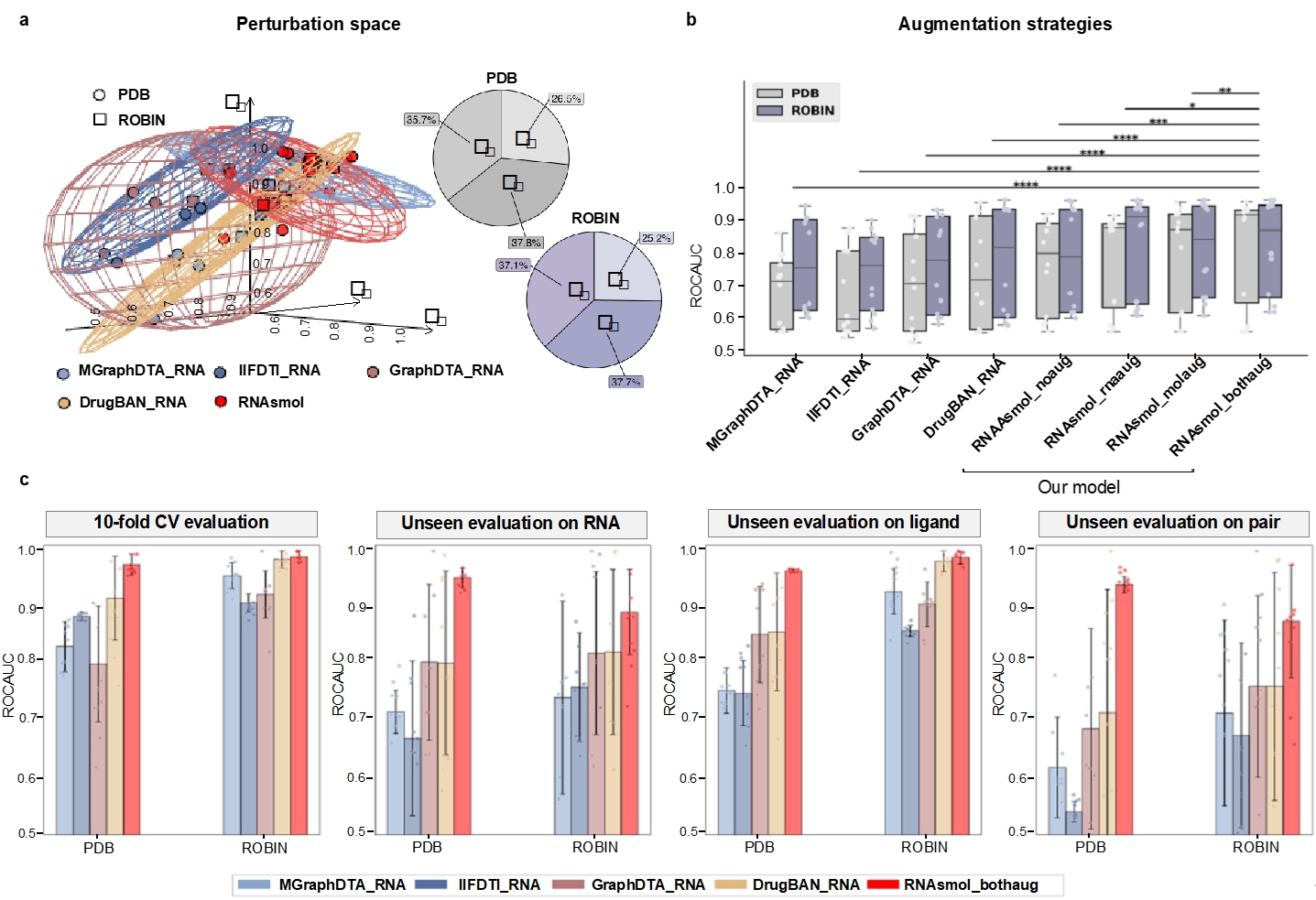
Figure 2. Evaluation of the prediction effect of RNAsmol on different validation sets
3. RNAsmol can accurately distinguish between bait molecules and real ligands as a virtual screening tool
In the application of virtual screening, RNAsmol shows unique advantages. Unlike traditional screening methods that rely on structural information, RNAsmol makes predictions based entirely on RNA sequence information. Because the three-dimensional structural data of many disease-related RNA targets (such as lncRNA) are often difficult to obtain, RNAsmol can fill this data gap to achieve predictive screening of these targets. Experimental results show that RNAsmol successfully improved the ranking score by about 30% when distinguishing between bait molecules and real ligands. Therefore, RNAsmol has a wide range of applicability in various RNA-targeted drug screening, and this method can be used to screen potential drug molecules more efficiently.
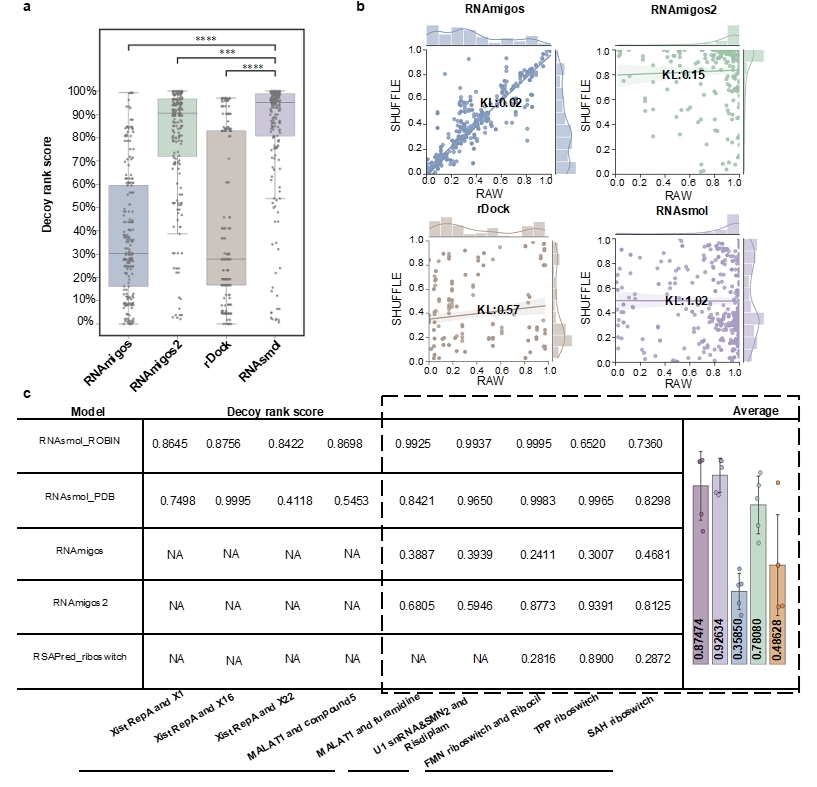
Figure 3 Comparison of RNAsmol and other computational methods
In general, this study provides new ideas for computational modeling of targeted RNA drug development by exploring the application of deep learning training strategies based on data perturbation and augmentation in data-scarce scenarios.
Professor Zhi John Lu from the School of Life Sciences of Tsinghua University and Professor Zhenjiang Xu from Nanchang University are the corresponding authors of the paper. Hongli Ma, a former postdoctoral fellow at Tsinghua University, is the first author of the article. This project was funded by the National Key R&D Program of China, the National Natural Science Foundation of China, the Chinese Ministry of Education Key Laboratory of Bioinformatics, the National Key Laboratory of Green Biomanufacturing of China, the Institute of Precision Medicine of Tsinghua University, and Bayer Pharmaceuticals.
Link to the paper:https://www.nature.com/articles/s43588-025-00820-x
Editor: Li Han

